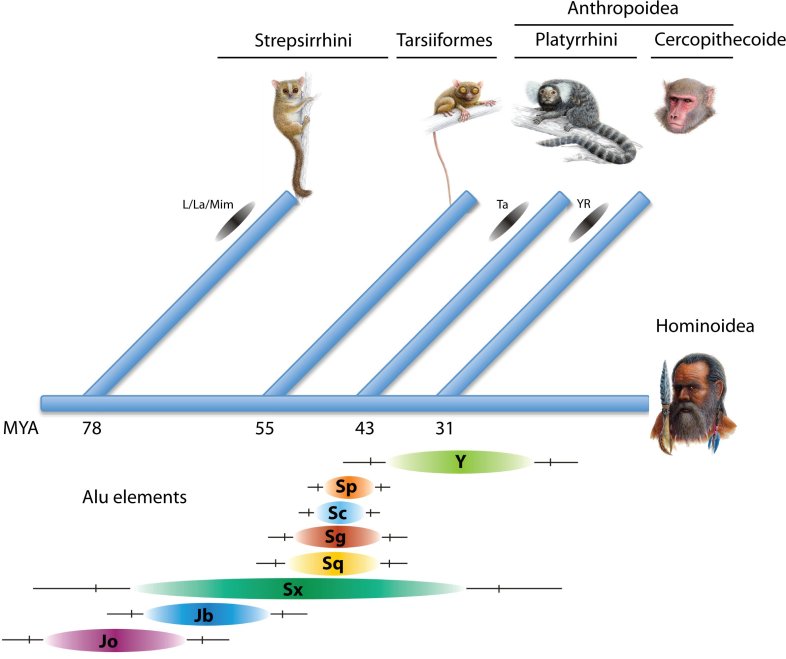
Considering the high frequency of transposed elements (e.g., mammalian genomes are composed of nearly 50% transposed elements) transposed elements contribute significantly to genome evolution and encrypt valuable information about the phylogenetic relationships of their host species.
The principle behind the application is that younger active transposed elements are able to insert into older elements but not vice versa. Thus, having old and young inserted elements as coexisting, nested components of current genomes provides retrospective information about their historical activity.
Using RepeatMasker reports as a source for TinT analyses, predicted nested insertions are evaluated, sorted, and introduced into a mathematical probabilistic model to trace their probable activity periods.
- Input: RepeatMasker outfiles. Predefined outfiles are available in a selection box.
- Parameters: Selecting and merging transposed elements of interest
- Variable: Stringency of detecting nested elements in RepeatMasker reports can be selected.
- Outfile: Matrix of inserted versus interrupted elements. Graph of probable activity (ovals represent 75%, vertical lines 95% and horizontal lines 99% of probable activity) Activity patterns are printable or exportable as postscript or png files.
Reference: Churakov G, Grundmann N, Kuritzin A, Brosius J, Makalowski W, Schmitz J. (2010) A Novel Web-Based TinT Application and the Chronology of the Primate Alu Retroposon Activity. BMC Evolutionary Biology 10:376. [Reprint]
The probilistic model used by Tint is described here
The primate paintings were provided by Jón Baldur Hliðberg